I am rather new to jupyter notebook, although I worked quite a bit with the 32-bit version of Enthought on a Debian Linux 32-bit machine. Then I switched to a newer 64-bit machine with Debian 9 64-bit. And the canopy-version of Enthought did not work.
Because RTP recommended it: I went to anaconda … 
Now I am playing with the notebook issues and found some strange effects, part of them having been solved by this discourse platform like implementing the environment variable PYTHONPATH.
So much of my problems seem to stem from not understanding the pertaining documentation (I downloaded a ~100 page pdf-file) …
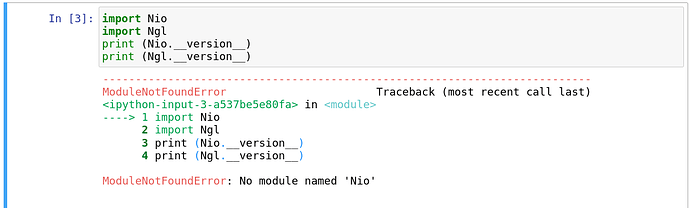
Now within the modules to be imported to notebook I had to install Nio and Ngl.
I did this:
(base) joerg@primergy:~$ conda create --name pyn_env --channel conda-forge pynio pyngl
and got the following results on a terminal-console:
lot of stuff which was ok anyway
next I did:
(base) joerg@primergy:~ conda activate pyn_env*
*(pyn_env) joerg@primergy:~ python
*Python 3.8.6 | packaged by conda-forge | (default, Dec 26 2020, 05:05:16) *
[GCC 9.3.0] on linux
Type “help”, “copyright”, “credits” or “license” for more information.
>>> import Nio
>>> import Ngl
print(Nio.version)?*
>>> print ( Nio.version)
1.5.5
>>> print ( Ngl.version)
1.6.1
>>>
so far so good,
Next step was to use Nio and Ngl within jupyter.
I started a jupyter machine - but it did not find the newly installed modules.
What did i forget to do within jupyter?
I forgot - after calling the conda command (above) I at the start of this command’s output I got the following:
Collecting package metadata (current_repodata.json): done
Solving environment: failed with repodata from current_repodata.json, will retry with next repodata source.
Collecting package metadata (repodata.json): done
Solving environment: done
## Package Plan ##
- environment location: /home/joerg/anaconda3/envs/pyn_env*
- added / updated specs:*
-
- pyngl*
-
- pynio*
The following packages will be downloaded:
- pynio*
[…]