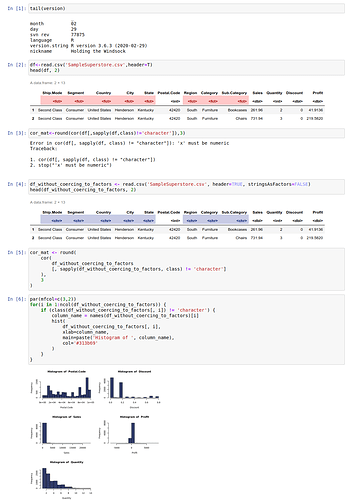
I was trying to find some correlations between different columns of a dataset and store them in a matrix to form a correlation matrix. For this, I wanted to use the cor function of R.
The link to the dataset: https://bit.ly/3i4rbWl
What I have done till now:
df<-read.csv('SampleSuperstore.csv',header=T) #Reading the dataset
df<-df[!duplicated(df),] #removing duplicate entries
Now as I go on to apply the cor function like this:
cor_mat<-round(cor(df[,sapply(df,class)!='character']),3), Binder throws an error:
Error in cor(df[, sapply(df, class) != “character”]): ‘x’ must be numeric
Traceback:
- cor(df[, sapply(df, class) != “character”])
- stop(“‘x’ must be numeric”)
But, in my local machine, this same command with this same set of data and same conditions works flawlessly:
Not only this, I used Google Colaboratory to run my IPYNB file, and there also, no errors came up.
Further, for my work, I had to plot some histograms of this dataset and I went on to do those as follows:
par(mfcol=c(3,2))
for(i in 1:ncol(df))
{
if (class(df[,i])!='character')
{
hist(df[,i],xlab=names(df)[i],main=paste('Histogram of ',
names(df)[i]),col='#313b69')
}
}
Here also, Binder threw an error:
Error in hist.default(df[, i], xlab = names(df)[i], main = paste("Histogram of ", : ‘x’ must be numeric
Traceback:
- hist(df[, i], xlab = names(df)[i], main = paste("Histogram of ",
. names(df)[i]), col = “#313b69”)- hist.default(df[, i], xlab = names(df)[i], main = paste("Histogram of ",
. names(df)[i]), col = “#313b69”)- stop(“‘x’ must be numeric”)
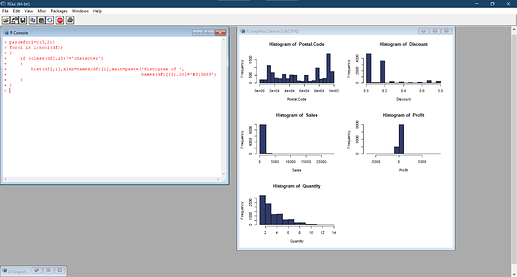
But in my local machine and Google Colaboratory, things worked flawlessly again:
Seeing the nature of the error, I found out that the variable should be numeric to be eligible to apply cor and hist. However, I provided a check for this criterion only and then applied the functions where these criteria are met. My local system and Google colab demonstrated that there is nothing wrong with my code. But Binder is showing else.
Can someone help me to understand why this discrepancy?
Edit 1: As I moved up one trust level, I can now put multiple images in my posts, thus, updating the post with the two images which I thought to be relevant

 The 4.0 release of R has brought some long requested changes, including a change to the default value of the
The 4.0 release of R has brought some long requested changes, including a change to the default value of the